full-FORCE and “Classic FORCE” learning with spikes¶
This notebook demonstrates how to implement both full-FORCE [1] and “Classic FORCE” [2] networks in Nengo. This makes it “trivial” to switch between neuron models (rate-based, spiking, adaptive, etc.), and to explore the effects of different learning rules and architectural assumptions.
For this demonstration, we use recursive least-squares (RLS) learning, with spiking LIF neurons, and the two basic architectures (full-FORCE and classic-FORCE) – to learn a bandpass filter (a.k.a. a “decaying oscillator” triggered by unit impulses).
[1]:
%pylab inline
import pylab
try:
import seaborn as sns # optional; prettier graphs
except ImportError:
pass
import numpy as np
import nengo
import nengolib
from nengolib import RLS, Network
Populating the interactive namespace from numpy and matplotlib
[2]:
# Task parameters
pulse_interval = 1.0
amplitude = 0.1
freq = 3.0
decay = 2.0
dt = 0.002
trials_train = 3
trials_test = 2
# Fixed model parameters
n = 200
seed = 0
rng = np.random.RandomState(seed)
ens_kwargs = dict( # neuron parameters
n_neurons=n,
dimensions=1,
neuron_type=nengo.LIF(), # nengolib.neurons.Tanh()
intercepts=[-1]*n, # intercepts are irelevant for Tanh
seed=seed,
)
# Hyper-parameters
tau = 0.1 # lowpass time-constant (10ms in [1])
tau_learn = 0.1 # filter for error / learning (needed for spiking)
tau_probe = 0.05 # filter for readout (needed for spiking
learning_rate = 0.1 # 1 in [1]
g = 1.5 / 400 # 1.5 in [1], scaled by firing rates
g_in = tau / amplitude # scale the input encoders (usually 1)
g_out = 1.0 # scale the recurrent encoders (usually 1)
# Pre-computed constants
T_train = trials_train * pulse_interval
T_total = (trials_train + trials_test) * pulse_interval
[3]:
with Network(seed=seed) as model:
# Input is a pulse every pulse_interval seconds
U = np.zeros(int(pulse_interval / dt))
U[0] = amplitude / dt
u = nengo.Node(output=nengo.processes.PresentInput(U, dt))
# Desired output is a decaying oscillator
z = nengo.Node(size_in=1)
nengo.Connection(u, z, synapse=nengolib.synapses.Bandpass(freq, decay))
# Initial weights
e_in = g_in * rng.uniform(-1, +1, (n, 1)) # fixed encoders for f_in (u_in)
e_out = g_out * rng.uniform(-1, +1, (n, 1)) # fixed encoders for f_out (u)
JD = rng.randn(n, n) * g / np.sqrt(n) # target-generating weights (variance g^2/n)
Classic FORCE¶
xCare the neuronssCare the unfiltered currents into each neuron (sC -> Lowpass(tau) -> xC)zCis the learned output estimate, decoded by the neurons, and re-encoded back intosCalongside some random feedback (JD)eCis a gated error signal for RLS that turns off afterT_trainseconds. This error signal learns the feedback decoders by minmizing the difference betweenz(ideal output) andzC(actual output).
The error signal driving RLS has an additional filter applied (tau_learn) to handle the case when this signal consists of spikes (not rates).
[4]:
with model:
xC = nengo.Ensemble(**ens_kwargs)
sC = nengo.Node(size_in=n) # pre filter
eC = nengo.Node(size_in=1, output=lambda t, e: e if t < T_train else 0)
zC = nengo.Node(size_in=1) # learned output
nengo.Connection(u, sC, synapse=None, transform=e_in)
nengo.Connection(sC, xC.neurons, synapse=tau)
nengo.Connection(xC.neurons, sC, synapse=None, transform=JD) # chaos
connC = nengo.Connection(
xC.neurons, zC, synapse=None, transform=np.zeros((1, n)),
learning_rule_type=RLS(learning_rate=learning_rate, pre_synapse=tau_learn))
nengo.Connection(zC, sC, synapse=None, transform=e_out)
nengo.Connection(zC, eC, synapse=None) # actual
nengo.Connection(z, eC, synapse=None, transform=-1) # ideal
nengo.Connection(eC, connC.learning_rule, synapse=tau_learn)
full-FORCE¶
Figure 1. Network architecture from [1].
Target-Generating Network¶
See Fig 1b.
xDare the neurons that behave like classic FORCE in the ideal case (assuming the ideal outputzis perfectly re-encoded)sDare the unfiltered currents into each neuron (sD -> Lowpass(tau) -> xD)
[5]:
with model:
xD = nengo.Ensemble(**ens_kwargs)
sD = nengo.Node(size_in=n) # pre filter
nengo.Connection(u, sD, synapse=None, transform=e_in)
nengo.Connection(z, sD, synapse=None, transform=e_out)
nengo.Connection(sD, xD.neurons, synapse=tau)
nengo.Connection(xD.neurons, sD, synapse=None, transform=JD)
Task-Performing Network¶
See Fig 1a.
xFare the neuronssFare the unfiltered currents into each neuron (sF -> Lowpass(tau) -> xF)eFis a gated error signal for RLS that turns off afterT_trainseconds. This error signal learns the full-rank feedback weights by minimizing the difference between the unfiltered currentssDandsF.
The error signal driving RLS also has the same filter applied (tau_learn) to handle spikes. The output estimate is trained offline from the entire training set using batched least-squares, since this gives the best performance.
[6]:
with model:
xF = nengo.Ensemble(**ens_kwargs)
sF = nengo.Node(size_in=n) # pre filter
eF = nengo.Node(size_in=n, output=lambda t, e: e if t < T_train else np.zeros_like(e))
nengo.Connection(u, sF, synapse=None, transform=e_in)
nengo.Connection(sF, xF.neurons, synapse=tau)
connF = nengo.Connection(
xF.neurons, sF, synapse=None, transform=np.zeros((n, n)),
learning_rule_type=RLS(learning_rate=learning_rate, pre_synapse=tau_learn))
nengo.Connection(sF, eF, synapse=None) # actual
nengo.Connection(sD, eF, synapse=None, transform=-1) # ideal
nengo.Connection(eF, connF.learning_rule, synapse=tau_learn)
Results¶
[7]:
with model:
# Probes
p_z = nengo.Probe(z, synapse=tau_probe)
p_zC = nengo.Probe(zC, synapse=tau_probe)
p_xF = nengo.Probe(xF.neurons, synapse=tau_probe)
with nengo.Simulator(model, dt=dt) as sim:
sim.run(T_total)
/home/arvoelke/anaconda3/envs/py36/lib/python3.6/site-packages/nengo/utils/numpy.py:79: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
v = a[inds]
/home/arvoelke/CTN/nengolib/nengolib/signal/system.py:198: UserWarning: y0 (None!=0) does not properly initialize the system; see Nengo issue #1124.
"Nengo issue #1124." % y0, UserWarning)
/home/arvoelke/CTN/nengolib/nengolib/signal/system.py:198: UserWarning: y0 (None!=0) does not properly initialize the system; see Nengo issue #1124.
"Nengo issue #1124." % y0, UserWarning)
/home/arvoelke/CTN/nengolib/nengolib/signal/system.py:198: UserWarning: y0 (None!=0) does not properly initialize the system; see Nengo issue #1124.
"Nengo issue #1124." % y0, UserWarning)
[8]:
# We do the readout training for full-FORCE offline, since this gives better
# performance without affecting anything else
t_train = sim.trange() < T_train
t_test = sim.trange() >= T_train
solver = nengo.solvers.LstsqL2(reg=1e-2)
wF, _ = solver(sim.data[p_xF][t_train], sim.data[p_z][t_train])
zF = sim.data[p_xF].dot(wF)
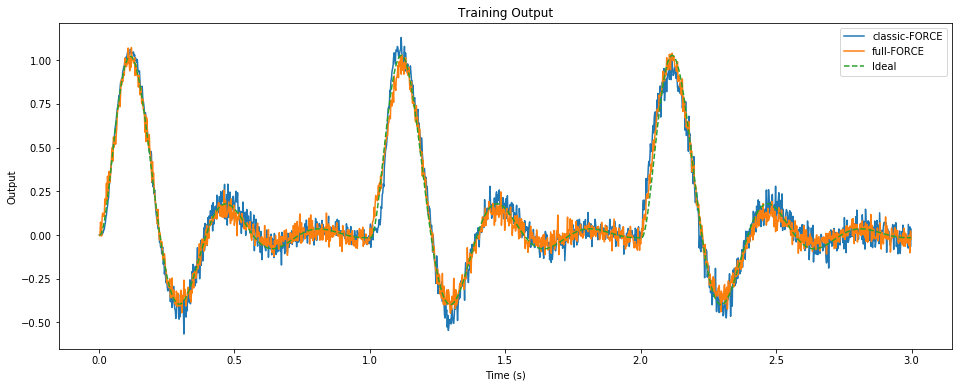
pylab.figure(figsize=(16, 6))
pylab.title("Training Output")
pylab.plot(sim.trange()[t_train], sim.data[p_zC][t_train], label="classic-FORCE")
pylab.plot(sim.trange()[t_train], zF[t_train], label="full-FORCE")
pylab.plot(sim.trange()[t_train], sim.data[p_z][t_train], label="Ideal", linestyle='--')
pylab.xlabel("Time (s)")
pylab.ylabel("Output")
pylab.legend()
pylab.show()
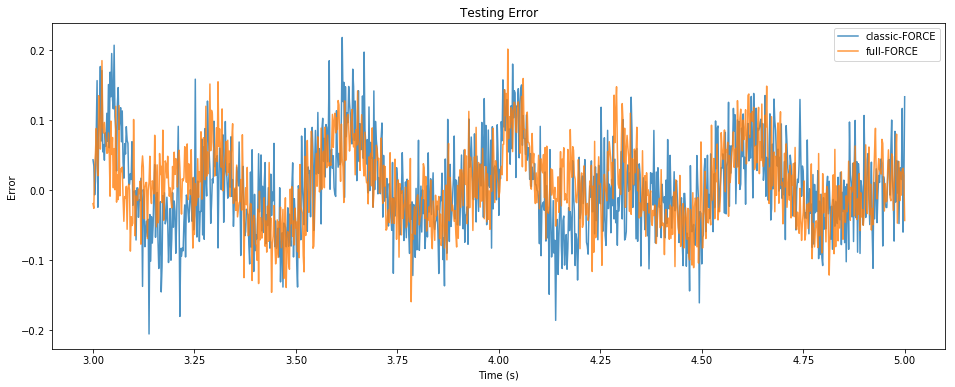
pylab.figure(figsize=(16, 6))
pylab.title("Testing Error")
pylab.plot(sim.trange()[t_test], sim.data[p_zC][t_test] - sim.data[p_z][t_test],
alpha=0.8, label="classic-FORCE")
pylab.plot(sim.trange()[t_test], zF[t_test] - sim.data[p_z][t_test],
alpha=0.8, label="full-FORCE")
pylab.xlabel("Time (s)")
pylab.ylabel("Error")
pylab.legend()
pylab.show()
References¶
[1] DePasquale, B., Cueva, C. J., Rajan, K., & Abbott, L. F. (2018). full-FORCE: A target-based method for training recurrent networks. PloS one, 13(2), e0191527. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0191527
[2] Sussillo, D., & Abbott, L. F. (2009). Generating coherent patterns of activity from chaotic neural networks. Neuron, 63(4), 544-557. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2756108/
[9]:
