Network Improvements¶
The class ``nengolib.Network()`` is intended to serve as a drop-in replacement for ``nengo.Network()``. This new class:
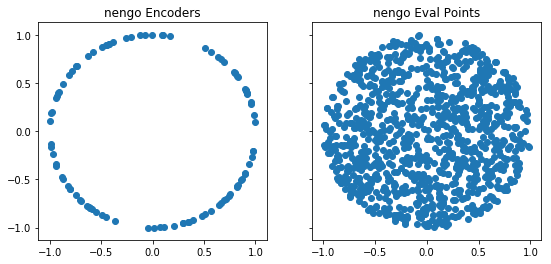
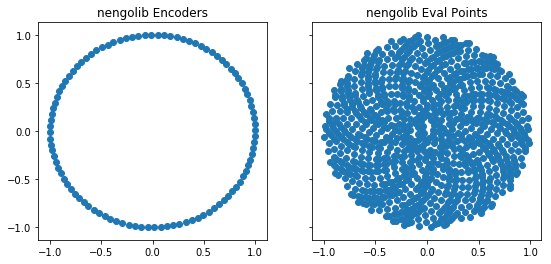
- samples
encodersmore uniformly; - samples
eval_pointsmore uniformly; and - uses neurons which spike at the ideal rate regardless of
dt(see Nengo #975; default innengo>=2.1.1).
As a result, the performance of ensembles should increase, both in terms of their representational quality (the encoders become better “representatives”) and the generalization error from the decoders (their approximation error on unseen test points). A current limitation is that the improvement only occurs for up to 40-dimensional ensembles (beyond this the original implementation is used).
[1]:
%pylab inline
import pylab
try:
import seaborn as sns # optional; prettier graphs
except ImportError:
pass
import nengo
import nengolib
def plot_points(module, n_neurons=100):
with module.Network() as model:
x = nengo.Ensemble(n_neurons, 2)
nengo.Connection(x, nengo.Node(size_in=2))
sim = nengo.Simulator(model)
fig, ax = pylab.subplots(1, 2, sharey=True, figsize=(9, 4))
ax[0].scatter(*sim.data[x].encoders.T)
ax[1].scatter(*sim.data[x].eval_points.T)
module = module.__name__.split('.', 1)[0]
ax[0].set_title('%s Encoders' % module)
ax[1].set_title('%s Eval Points' % module)
pylab.show()
plot_points(nengo)
plot_points(nengolib)
Populating the interactive namespace from numpy and matplotlib
0%
0%
[2]:
